Rhythmus is a new app that makes prescribing natural agents simpler than before when using the Opus23 database of natural agent effects on genes. Taking an overview of all SNPs for the client, the main Rhythmus screen shows a list of genes sorted by combined SNP effect power factor, with […]
Category: Curation
A tour of Cluster, the new Opus template editor
Utopia Demonstration Video
Utopia: Spectrum, visual community organization
SPECTRUM: Visual community organization SPECTRUM provides a visual representation of the taxonomic data at both the genus and phylum levels. The app demonstrates the weight, influence and diversity of your client’s microbiome in two helpful display formats, each of which are clickable for a deeper look and easy report curation. […]
Utopia: Loam, a fertile soil
Announcing the launch of Utopia, the suite of apps within Opus 23 that analyzes and reports on sequential data from uBiome tests. uBiome is the world’s first sequencing-based clinical microbiome screening test, giving the user insight into the bacterial population of multiple body areas. Utopia recognizes all bacteria found by uBiome, but is […]
A Roster of Opus 23 Algorithms
This is a current list of the multi-snp algorithms currently available in Opus 23. A few titles repeat, for the simple reason that two different algorithms, using different snps or genes, can result in a similar conclusion. Algorithms can be quite complex: it is not uncommon for one algorithm to […]
Decoding 23andMe ‘i’ Numbers
23andMe currently reports over 600,000 SNPs in the genome explorer, which are analyzed by their custom 2014 v4 chip. The process used is genotyping, rather than sequencing. The former is cheaper and quicker, and targets specific parts of the genome that are known to have variants in some or many people; the latter […]
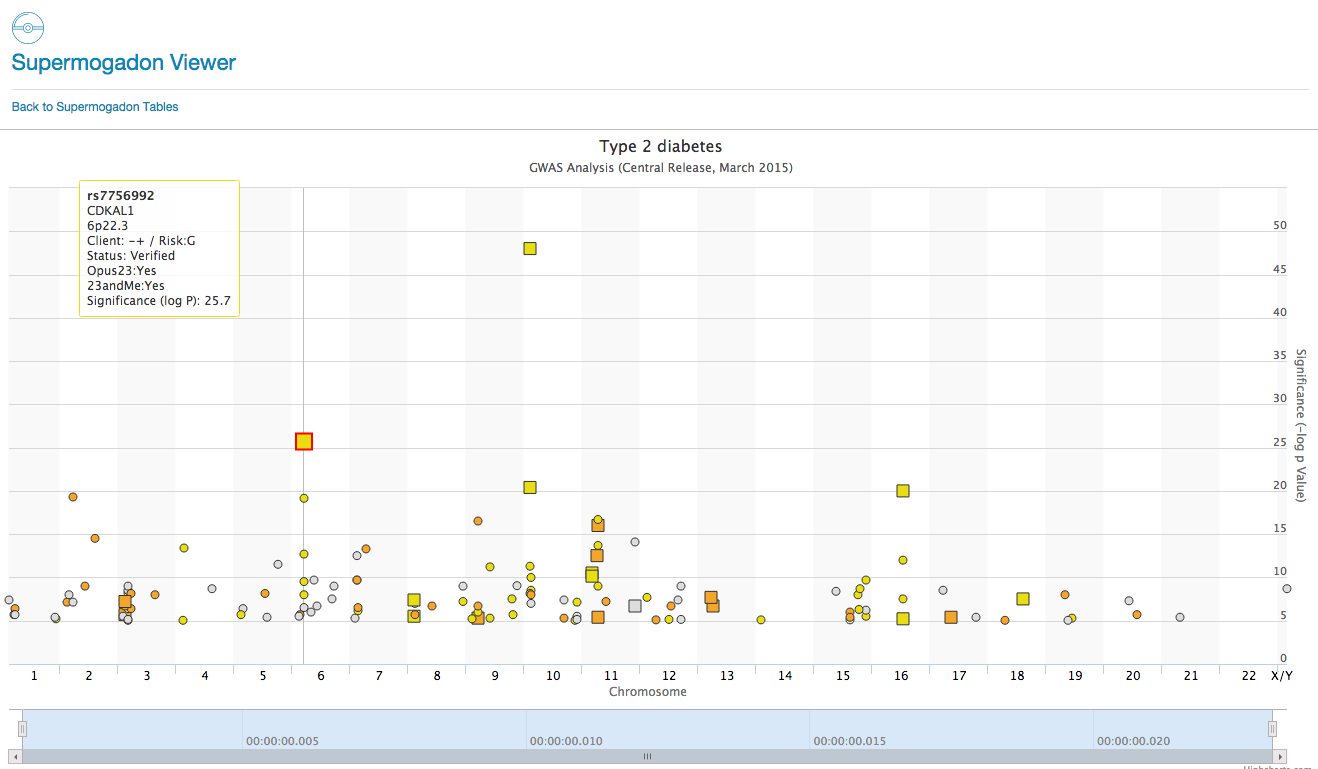
SuperMogadon
SuperMogadon is a highly flexible search and sort tool that allows you to easily compare the client’s genotype with results from Genome Wide Association Studies* (GWAS) through the Opus 23 Pro database. As you can see there are over 50+ pages of GWAS data in SuperMogadon, which would make grinding […]
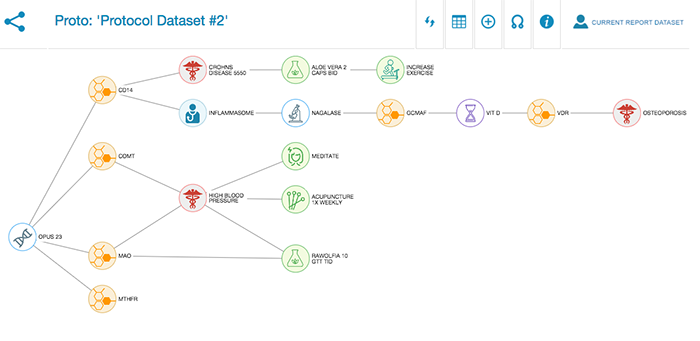
Protocol development
Two years ago I developed a software app called SkySaw for use on my teaching shift at the COEGM. SkySaw allows clinicians to structure patient encounters as a linked network (technically a directed acyclic graph). What made this attractive was that these individual networks could be connected together into a great […]
Client-friendly reporting
I designed Opus 23 Pro to serve two user audiences: the physician who works in the development environment to generate and curate information; and the client, who represents the end-user of that information. Both have widely differing needs and points of reference. One aspect of Opus 23 Pro that I […]