SPECTRUM: Visual community organization SPECTRUM provides a visual representation of the taxonomic data at both the genus and phylum levels. The app demonstrates the weight, influence and diversity of your client’s microbiome in two helpful display formats, each of which are clickable for a deeper look and easy report curation. […]
Tag: opus23
Utopia: Loam, a fertile soil
Announcing the launch of Utopia, the suite of apps within Opus 23 that analyzes and reports on sequential data from uBiome tests. uBiome is the world’s first sequencing-based clinical microbiome screening test, giving the user insight into the bacterial population of multiple body areas. Utopia recognizes all bacteria found by uBiome, but is […]
Lewis negative and secretor status
Many people are aware of the concept that their blood group influences their interaction with the environment: intestinal secretions of blood group antigens affect a person’s interaction with foods through being a marker of self recognised by the immune system. Similar to the way that a transfusion of blood from […]
Decoding 23andMe ‘i’ Numbers
23andMe currently reports over 600,000 SNPs in the genome explorer, which are analyzed by their custom 2014 v4 chip. The process used is genotyping, rather than sequencing. The former is cheaper and quicker, and targets specific parts of the genome that are known to have variants in some or many people; the latter […]
Psychic
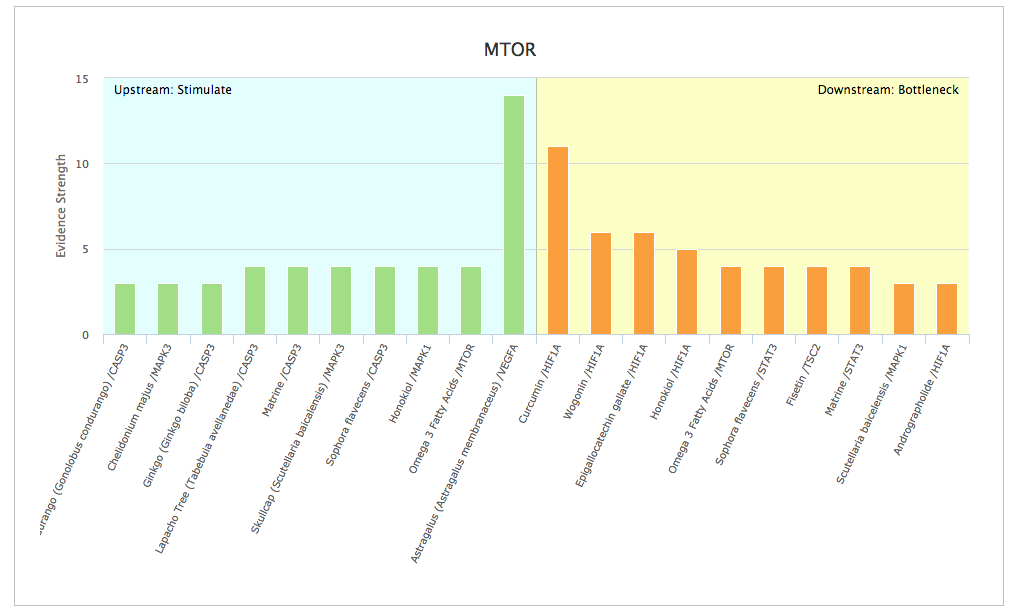
The Opus 23 PSYCHIC app allows you to search for natural products known to control gene expression. However, unlike a simple search engine, PSYCHIC is able to crawl up and down the molecular ‘Interactome’ (protein-protein interactions and gene expression data) to determine the upstream and downstream genes that interact with […]