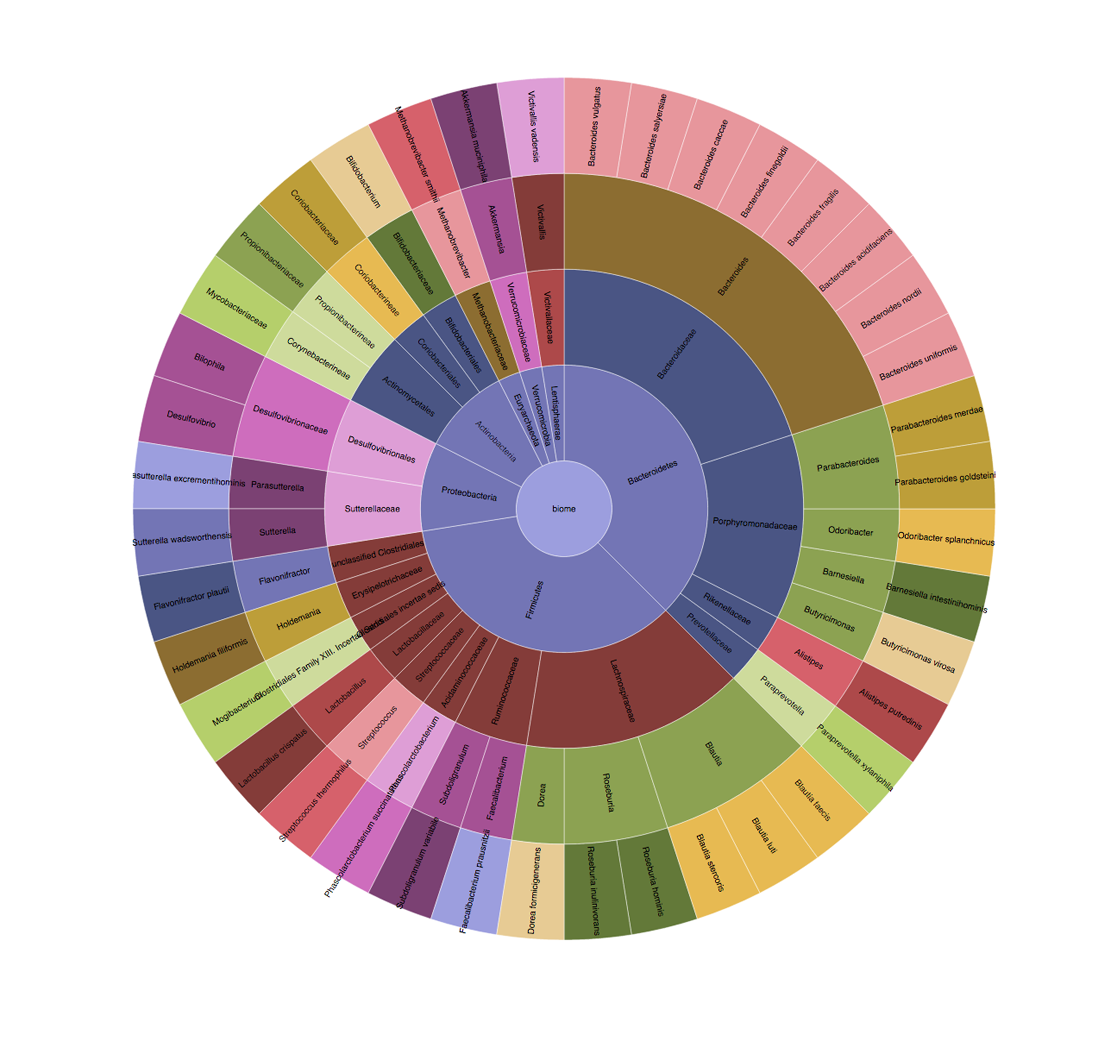
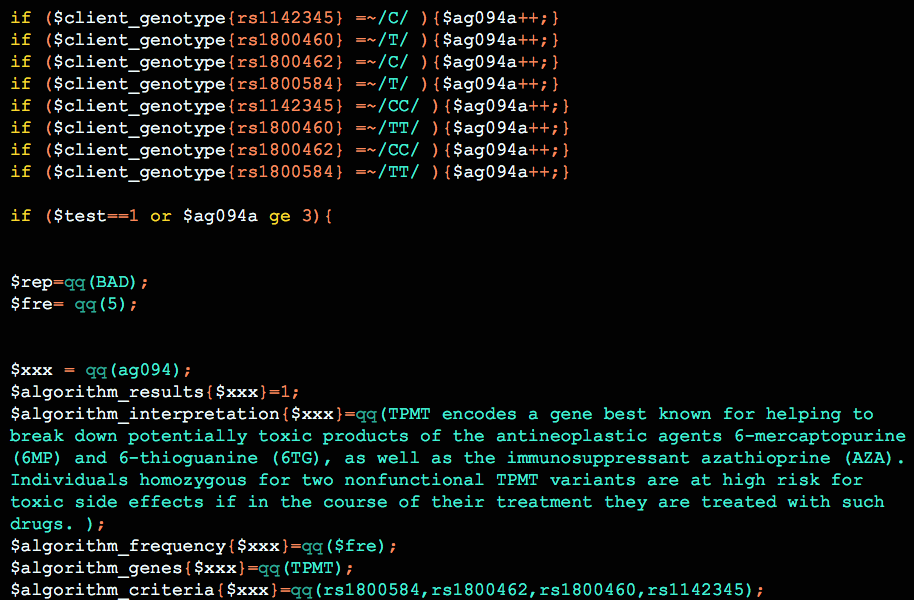
The Opus 23 genetic interpretation software [1] checks for ABH secretor status as part of the hereditary genetics algorithms. Knowing whether your client is a secretor or non-secretor is important for many reasons, one being that non-secretors have increased levels of serum vitamin B12. This is a well known association: at […]
Category: Coding/ Design
Opus 23 now supports multiple platforms
Decoding 23andMe ‘i’ Numbers
23andMe currently reports over 600,000 SNPs in the genome explorer, which are analyzed by their custom 2014 v4 chip. The process used is genotyping, rather than sequencing. The former is cheaper and quicker, and targets specific parts of the genome that are known to have variants in some or many people; the latter […]
Strobing the tissues
Psychic
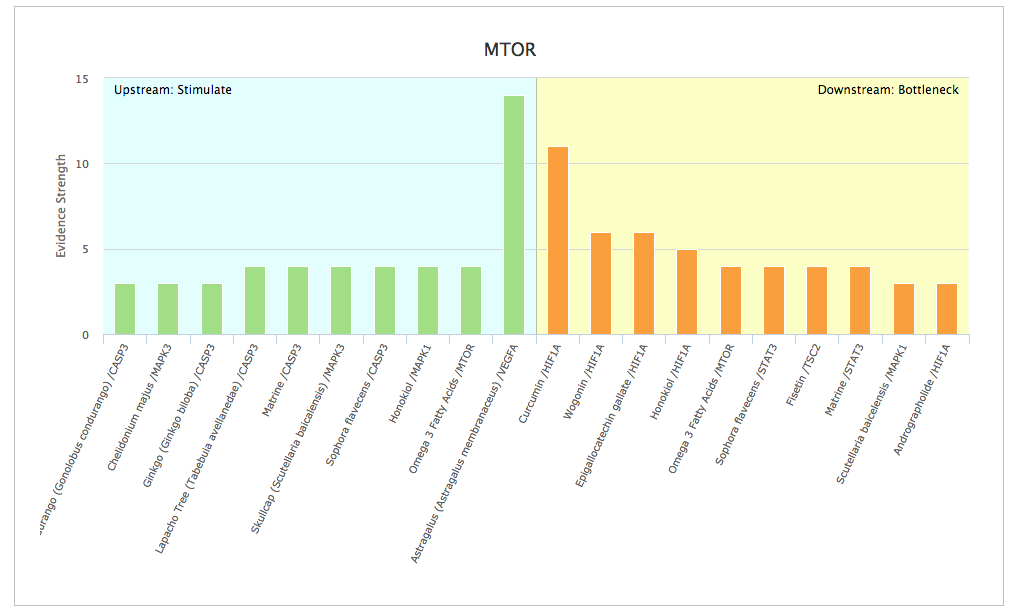
The Opus 23 PSYCHIC app allows you to search for natural products known to control gene expression. However, unlike a simple search engine, PSYCHIC is able to crawl up and down the molecular ‘Interactome’ (protein-protein interactions and gene expression data) to determine the upstream and downstream genes that interact with […]
Agency
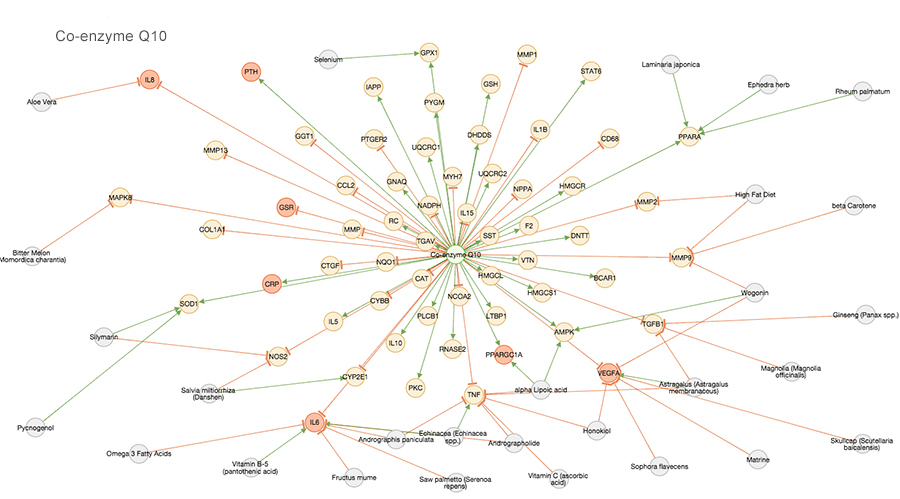
The Agency app in Opus 23 takes pharmacogenomics to the next level. Relying on the extensive Opus 23 database of published research detailing gene expression data linked to natural products, Agency provides a visual representation of their interactions web. Multiple agents can be displayed, allowing the clinician to synthesis multi-target […]
SuperMogadon
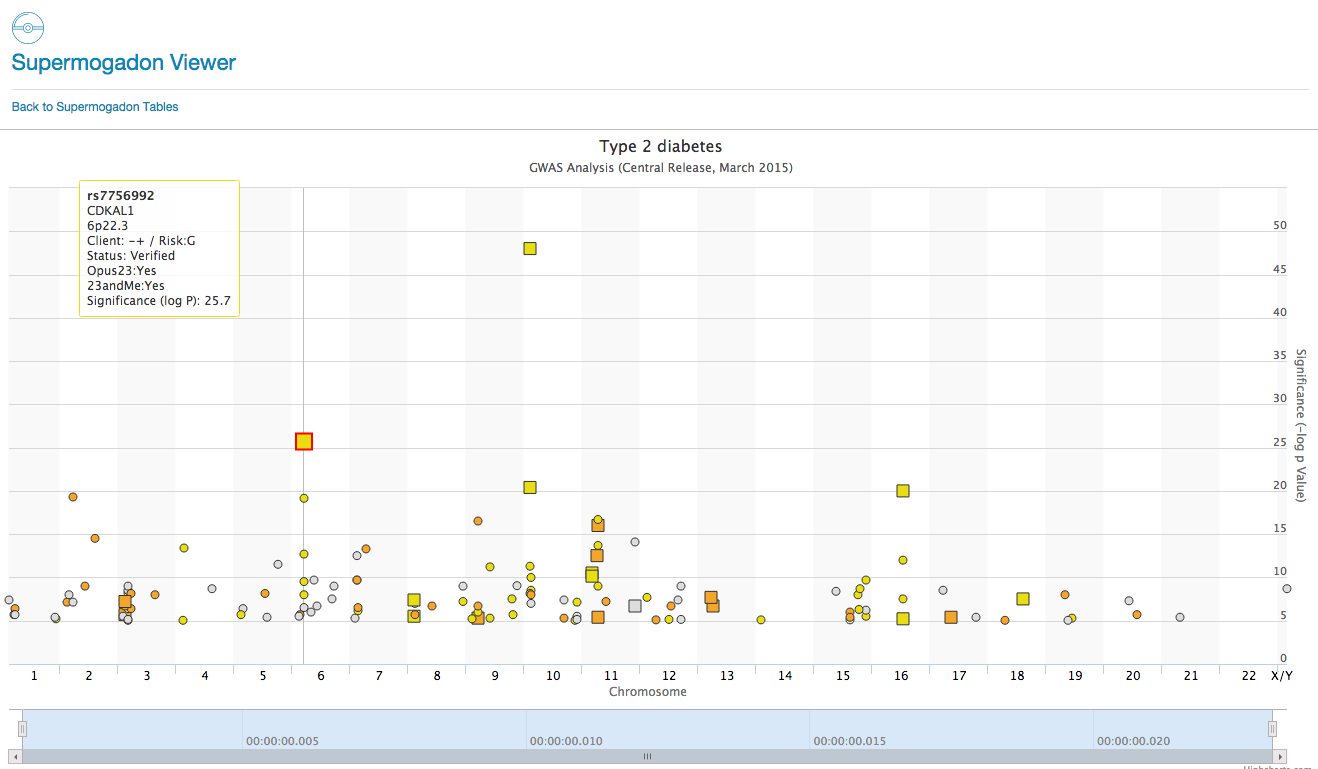
SuperMogadon is a highly flexible search and sort tool that allows you to easily compare the client’s genotype with results from Genome Wide Association Studies* (GWAS) through the Opus 23 Pro database. As you can see there are over 50+ pages of GWAS data in SuperMogadon, which would make grinding […]
Ancestry data is a no-go
Initially, I was very excited about the prospect of allowing the import of ancestry.com DNA data into Opus 23. After all, they use the same Illumina technology as 23andMe (although 23andMe apparently have their own unique chip.) Initial testing was promising. Like 23andMe, Ancestry supplies raw data in a basic […]