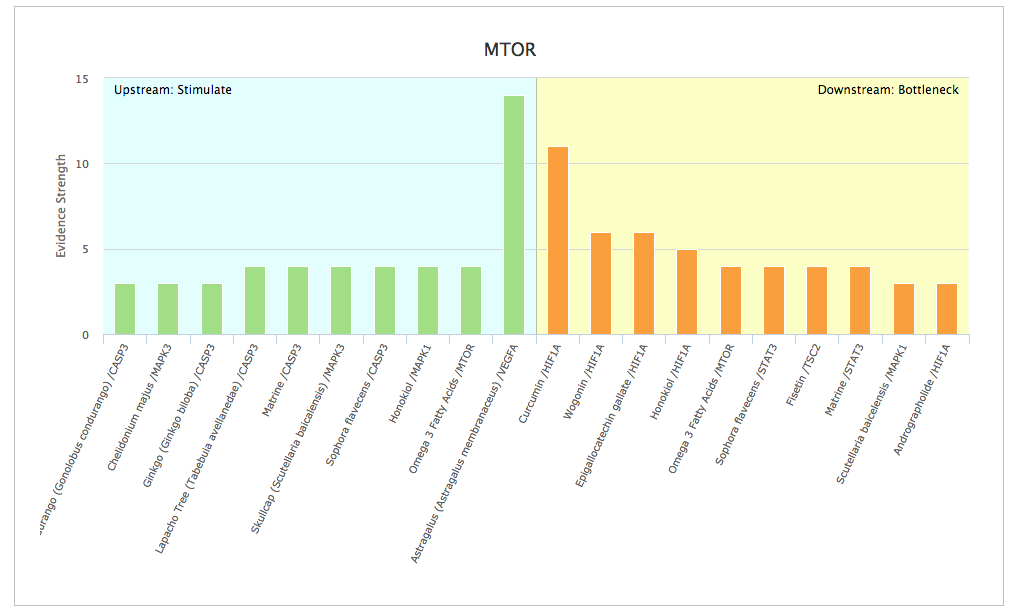
The Opus 23 PSYCHIC app allows you to search for natural products known to control gene expression. However, unlike a simple search engine, PSYCHIC is able to crawl up and down the molecular ‘Interactome’ (protein-protein interactions and gene expression data) to determine the upstream and downstream genes that interact with […]