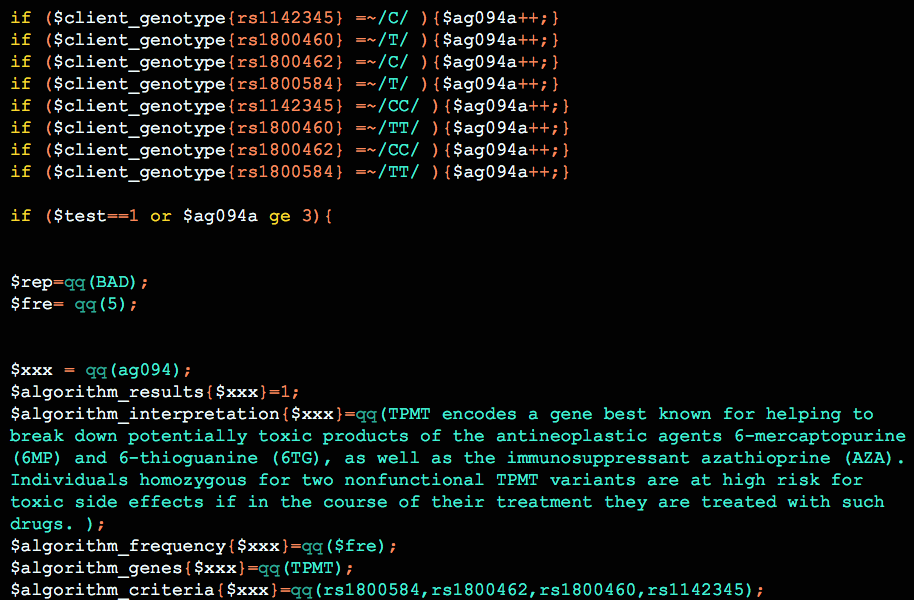
This is a current list of the multi-snp algorithms currently available in Opus 23. A few titles repeat, for the simple reason that two different algorithms, using different snps or genes, can result in a similar conclusion. Algorithms can be quite complex: it is not uncommon for one algorithm to […]
About: Peter
Posts by Peter:
Strobing the tissues
Psychic
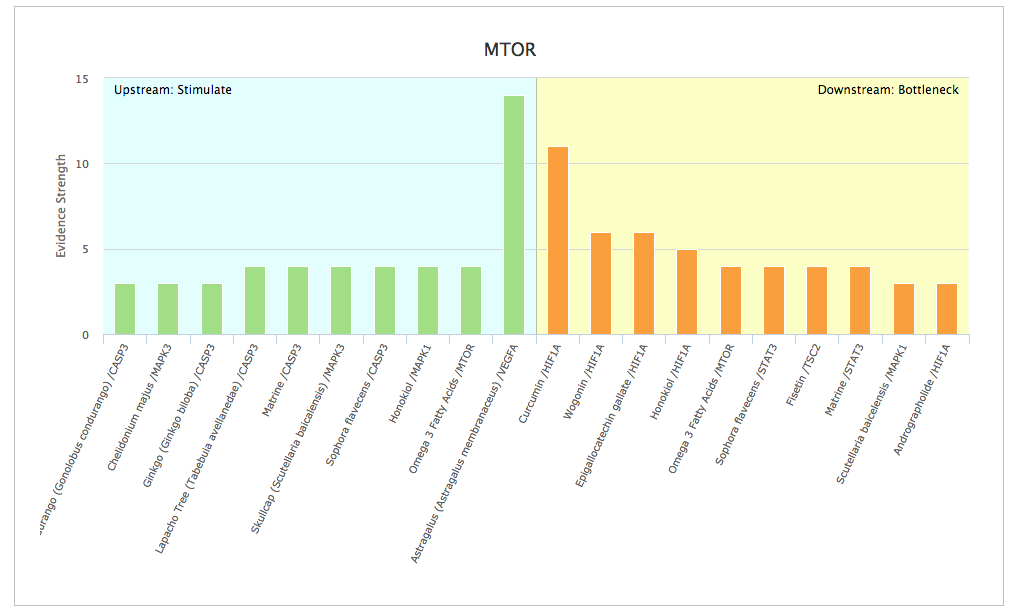
The Opus 23 PSYCHIC app allows you to search for natural products known to control gene expression. However, unlike a simple search engine, PSYCHIC is able to crawl up and down the molecular ‘Interactome’ (protein-protein interactions and gene expression data) to determine the upstream and downstream genes that interact with […]
Agency
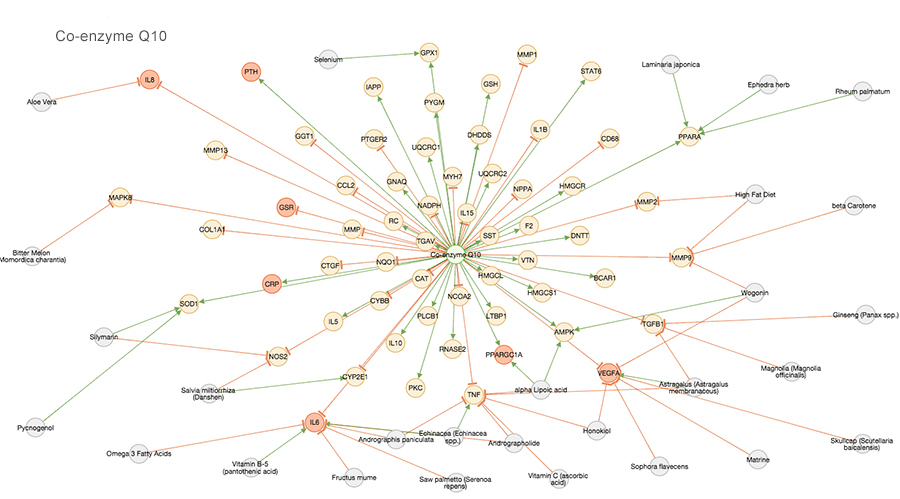
The Agency app in Opus 23 takes pharmacogenomics to the next level. Relying on the extensive Opus 23 database of published research detailing gene expression data linked to natural products, Agency provides a visual representation of their interactions web. Multiple agents can be displayed, allowing the clinician to synthesis multi-target […]
SuperMogadon
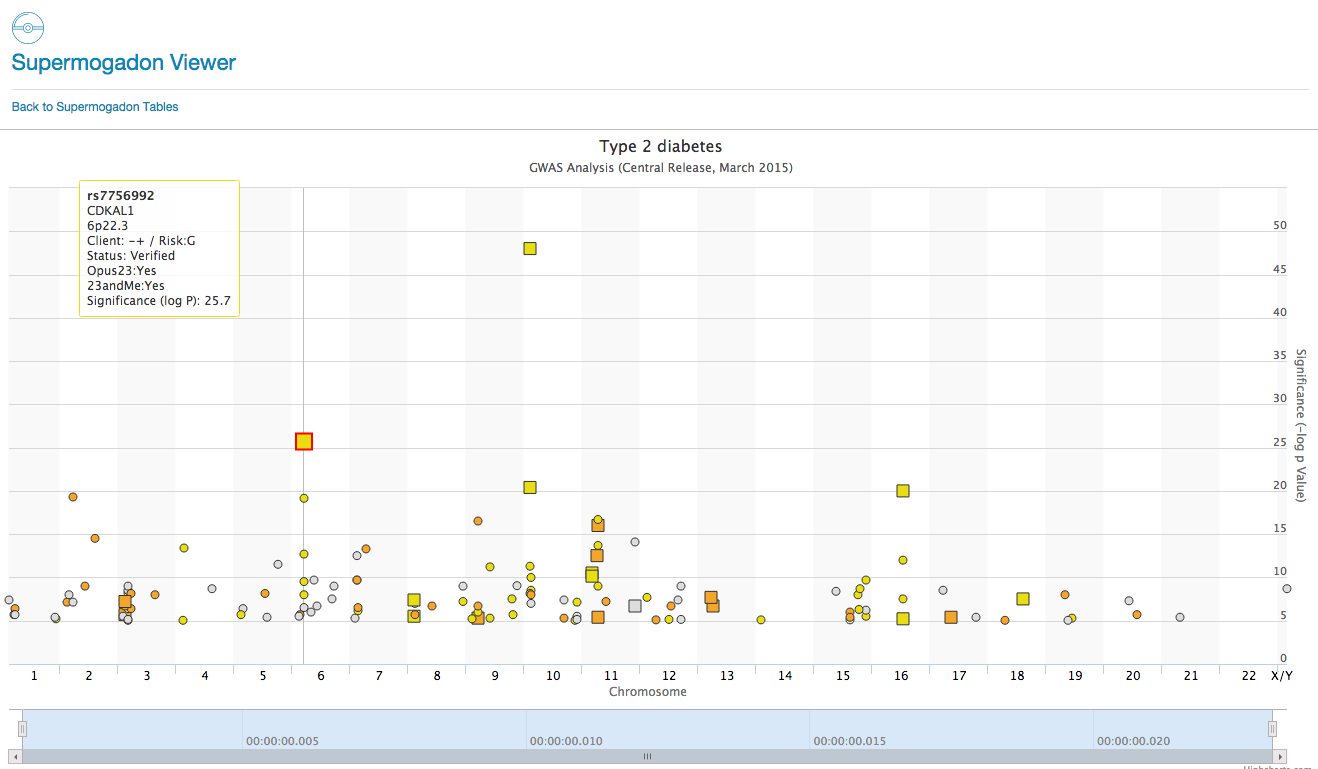
SuperMogadon is a highly flexible search and sort tool that allows you to easily compare the client’s genotype with results from Genome Wide Association Studies* (GWAS) through the Opus 23 Pro database. As you can see there are over 50+ pages of GWAS data in SuperMogadon, which would make grinding […]
MoboCaster
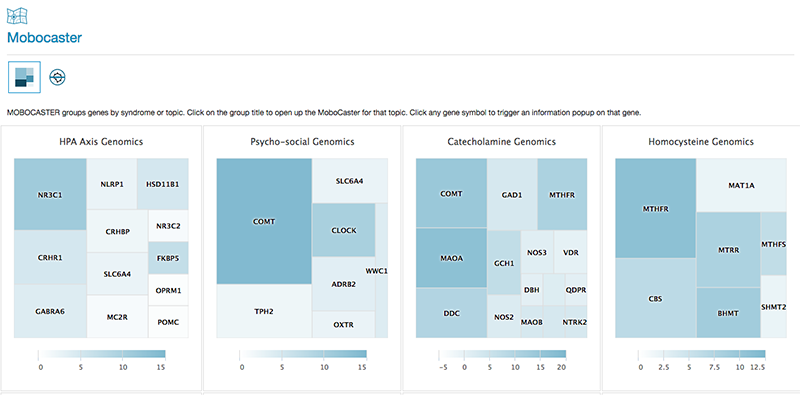
MoboCaster is an Opus 23 Pro informatics app that performs scenario-specific genomic analysis since, as clinicians, that’s pretty much how we think about genomics. The browser screen (above) in MoboCaster lists an overview of several genomic scenarios, such as the HPA Axis, Oxalate Genomics, Phase I Detoxification, etc. that display […]
Ancestry data is a no-go
Initially, I was very excited about the prospect of allowing the import of ancestry.com DNA data into Opus 23. After all, they use the same Illumina technology as 23andMe (although 23andMe apparently have their own unique chip.) Initial testing was promising. Like 23andMe, Ancestry supplies raw data in a basic […]
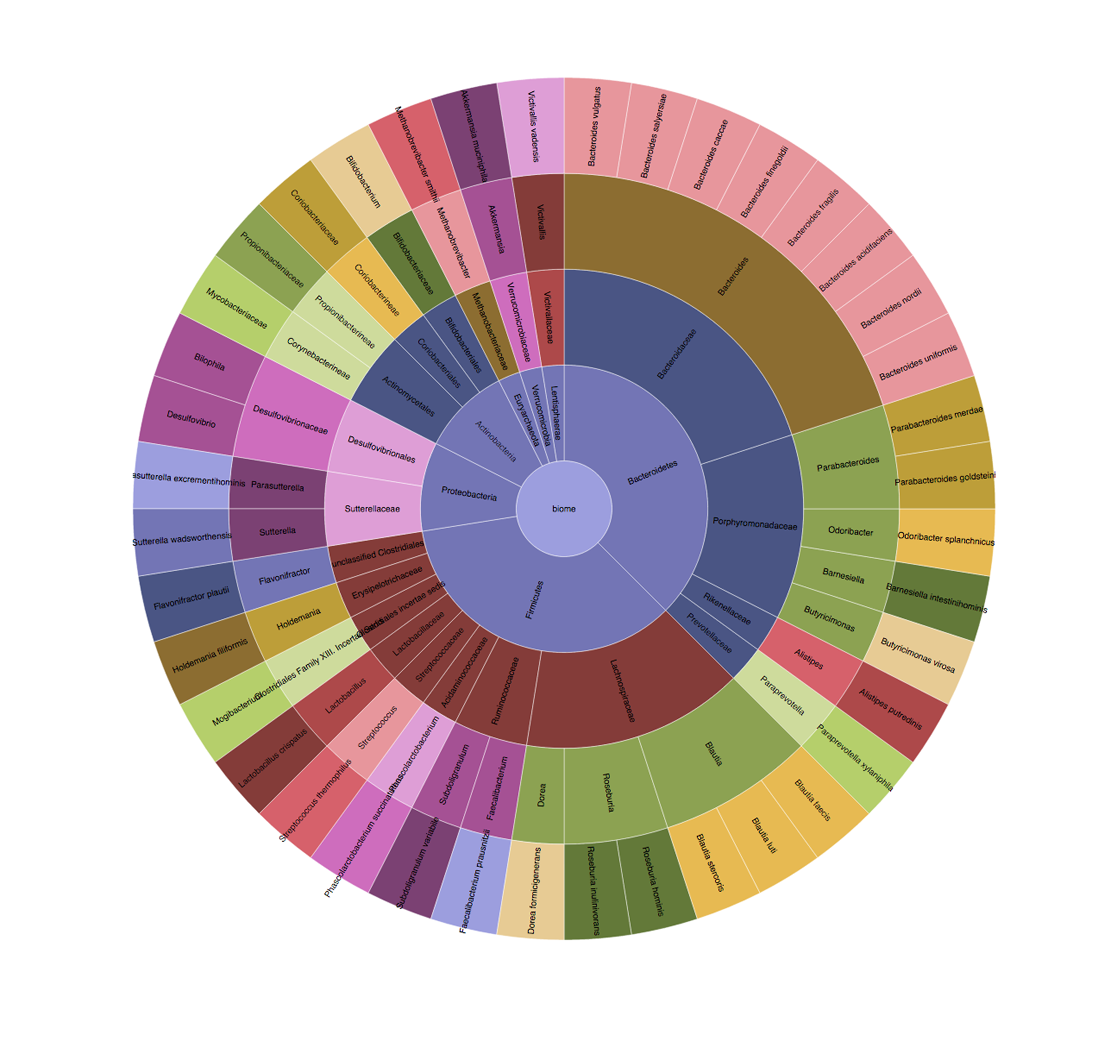
Microbiome mashup
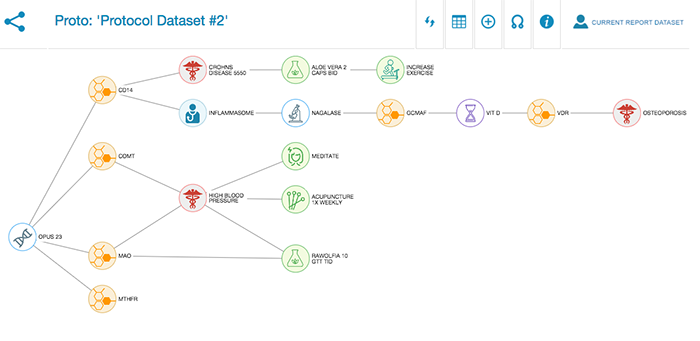
Protocol development
Two years ago I developed a software app called SkySaw for use on my teaching shift at the COEGM. SkySaw allows clinicians to structure patient encounters as a linked network (technically a directed acyclic graph). What made this attractive was that these individual networks could be connected together into a great […]