Rhythmus is a new app that makes prescribing natural agents simpler than before when using the Opus23 database of natural agent effects on genes. Taking an overview of all SNPs for the client, the main Rhythmus screen shows a list of genes sorted by combined SNP effect power factor, with […]
Category: Help Docs
Utopia: Advice, Intelligent interface with Opus23
Advice: Intelligent Interface with OPUS23 ADVICE is an app in Utopia that gives algorithmic considerations of multiple bacterial strains in conjunction with the client’s own genome to provide true/false clinical outcomes. ADVICE automatically pulls the client’s Opus23 genetic data where relevant, and incorporates it into algorithmic calculations. (comparable to LUMEN […]
Utopia: Pansophia, sequential outcome analysis
PANSOPHIA – sequential outcome analysis The genomic DNA analysis software Opus 23 includes the Utopia suite of apps for analyzing sequential uBiome data, comparing sequencing-based clinical microbiome data with the client’s own genomic DNA. PANSOPHIA is an Utopia app that gives you the power to sort your client’s data based upon associations with […]
Utopia: Spectrum, visual community organization
SPECTRUM: Visual community organization SPECTRUM provides a visual representation of the taxonomic data at both the genus and phylum levels. The app demonstrates the weight, influence and diversity of your client’s microbiome in two helpful display formats, each of which are clickable for a deeper look and easy report curation. […]
Utopia: Loam, a fertile soil
Announcing the launch of Utopia, the suite of apps within Opus 23 that analyzes and reports on sequential data from uBiome tests. uBiome is the world’s first sequencing-based clinical microbiome screening test, giving the user insight into the bacterial population of multiple body areas. Utopia recognizes all bacteria found by uBiome, but is […]
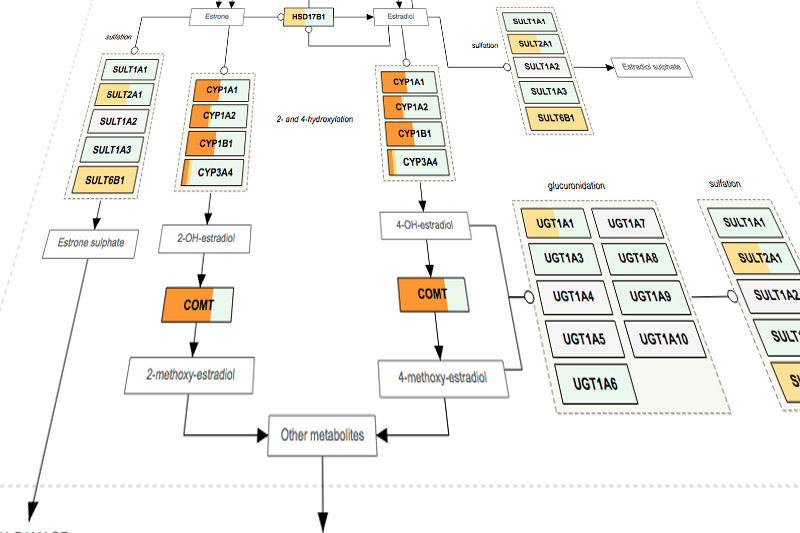
Don’t Fall into the Methyl Trap!
Methyl trapping is a situation in which folate becomes trapped and unusable by the body. It is defined as a functional folate deficiency that alters homocysteine metabolism such that folate–dependent resynthesis of methionine is compromised. Methyl trapping was originally described in Downs Syndrome (trisomy 21) where plasma levels of homocysteine, methionine, […]