Opus 23 provides many unique opportunities for data integration and visualization. One app that I’ve just been added to the Opus toolbox is STROBE, a new Opus analytic app that allows you drill-down client genomic data by organ, tissue or cell distribution. To do this Opus mashes up its own […]
Month: February 2016
Don’t Fall into the Methyl Trap!
Methyl trapping is a situation in which folate becomes trapped and unusable by the body. It is defined as a functional folate deficiency that alters homocysteine metabolism such that folate–dependent resynthesis of methionine is compromised. Methyl trapping was originally described in Downs Syndrome (trisomy 21) where plasma levels of homocysteine, methionine, […]
Psychic
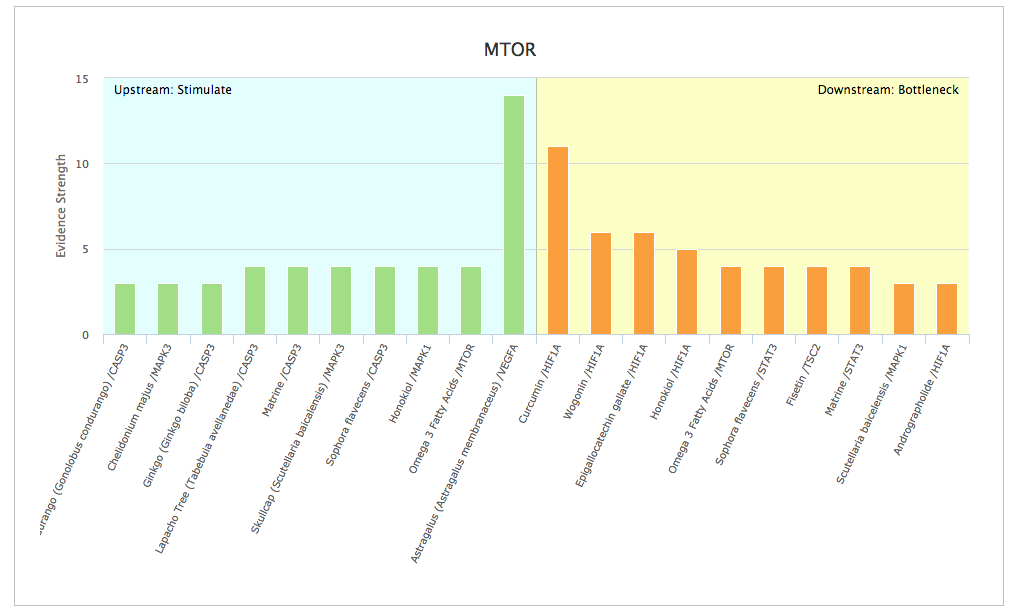
The Opus 23 PSYCHIC app allows you to search for natural products known to control gene expression. However, unlike a simple search engine, PSYCHIC is able to crawl up and down the molecular ‘Interactome’ (protein-protein interactions and gene expression data) to determine the upstream and downstream genes that interact with […]