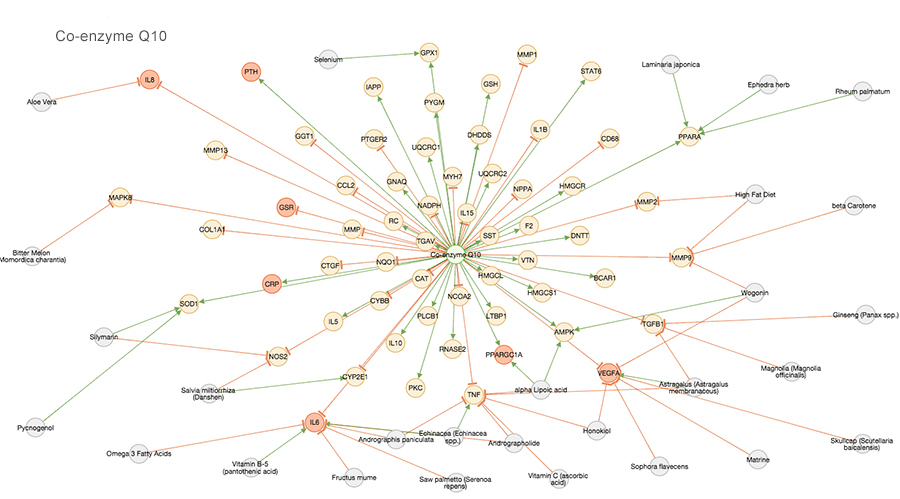
The Agency app in Opus 23 takes pharmacogenomics to the next level. Relying on the extensive Opus 23 database of published research detailing gene expression data linked to natural products, Agency provides a visual representation of their interactions web. Multiple agents can be displayed, allowing the clinician to synthesis multi-target […]
Month: January 2016
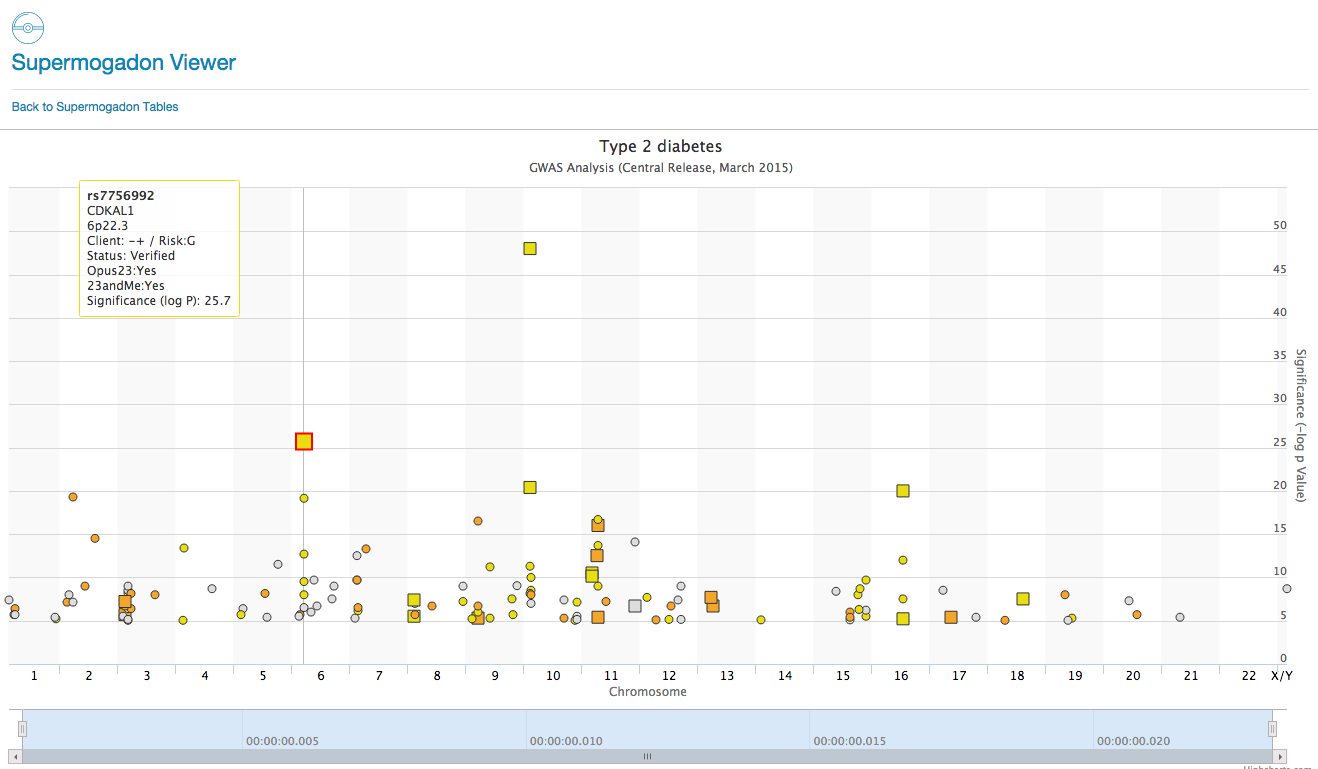
SuperMogadon
SuperMogadon is a highly flexible search and sort tool that allows you to easily compare the client’s genotype with results from Genome Wide Association Studies* (GWAS) through the Opus 23 Pro database. As you can see there are over 50+ pages of GWAS data in SuperMogadon, which would make grinding […]