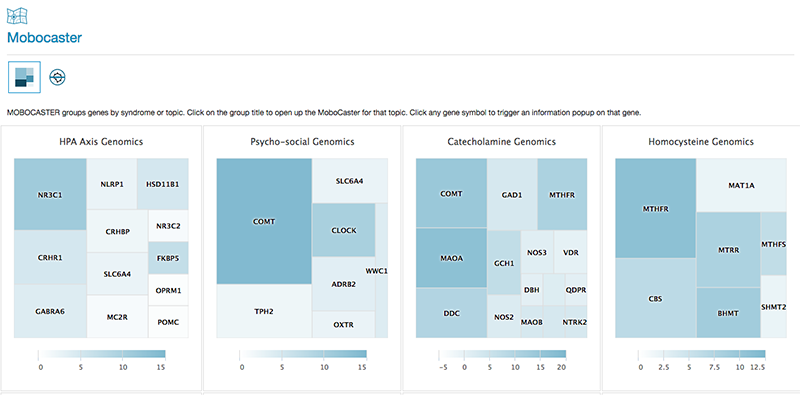
MoboCaster is an Opus 23 Pro informatics app that performs scenario-specific genomic analysis since, as clinicians, that’s pretty much how we think about genomics. The browser screen (above) in MoboCaster lists an overview of several genomic scenarios, such as the HPA Axis, Oxalate Genomics, Phase I Detoxification, etc. that display […]
Month: December 2015
Ancestry data is a no-go
Initially, I was very excited about the prospect of allowing the import of ancestry.com DNA data into Opus 23. After all, they use the same Illumina technology as 23andMe (although 23andMe apparently have their own unique chip.) Initial testing was promising. Like 23andMe, Ancestry supplies raw data in a basic […]
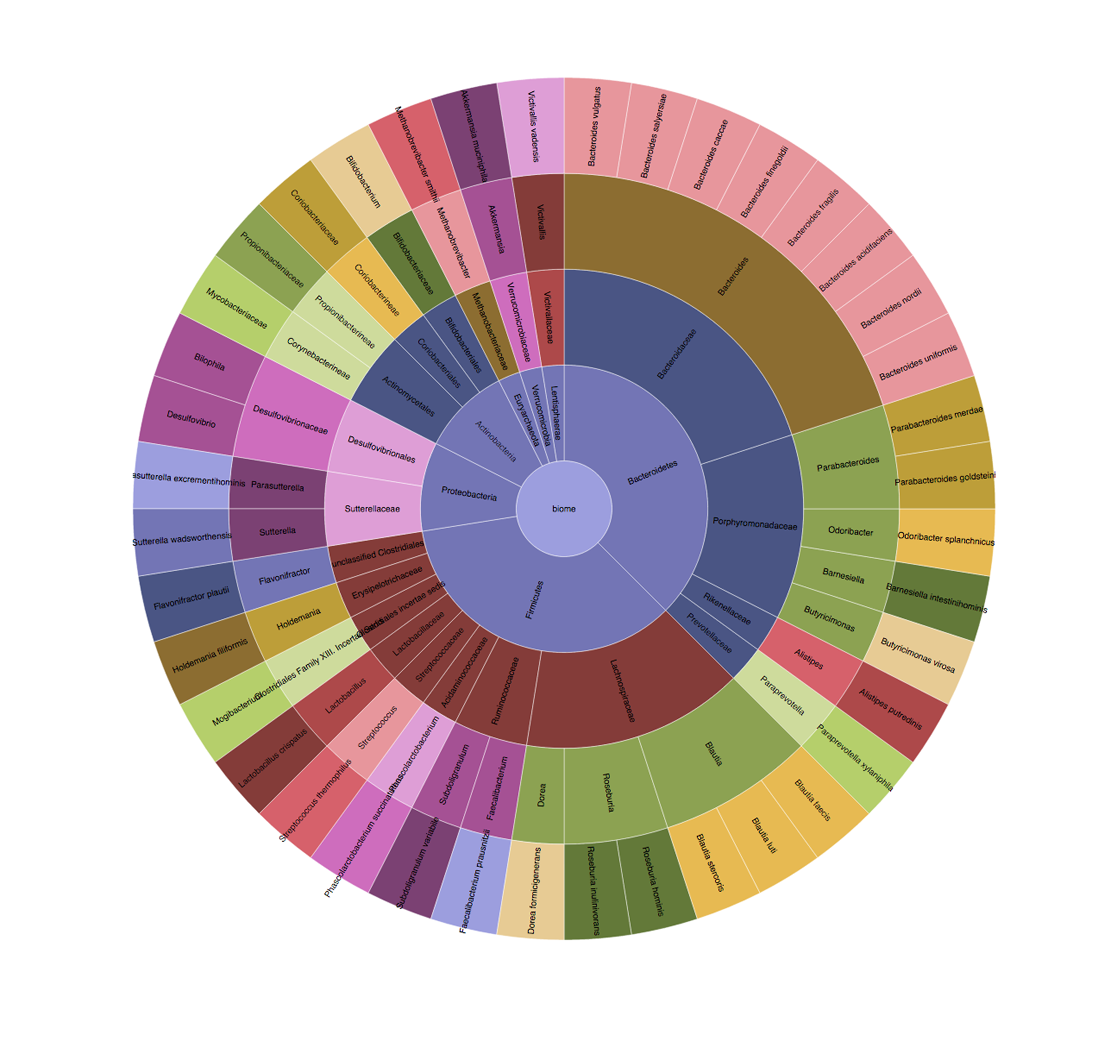
Microbiome mashup
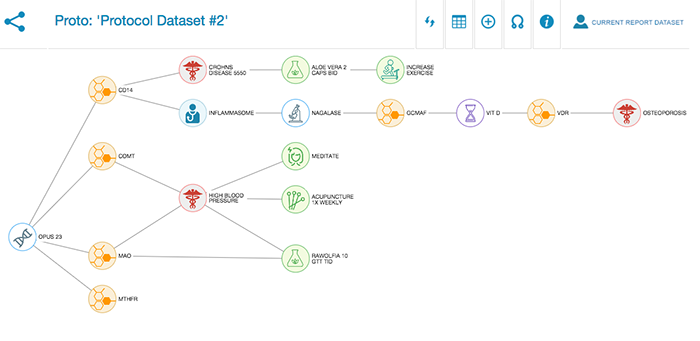
Protocol development
Two years ago I developed a software app called SkySaw for use on my teaching shift at the COEGM. SkySaw allows clinicians to structure patient encounters as a linked network (technically a directed acyclic graph). What made this attractive was that these individual networks could be connected together into a great […]
Pearl of an idea
Client-friendly reporting
I designed Opus 23 Pro to serve two user audiences: the physician who works in the development environment to generate and curate information; and the client, who represents the end-user of that information. Both have widely differing needs and points of reference. One aspect of Opus 23 Pro that I […]
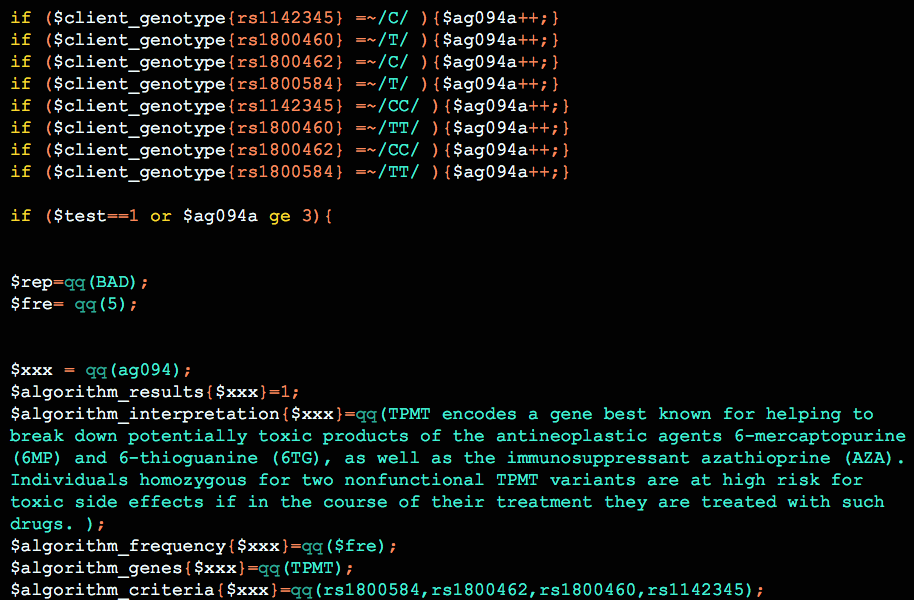
Multi-SNP algorithms
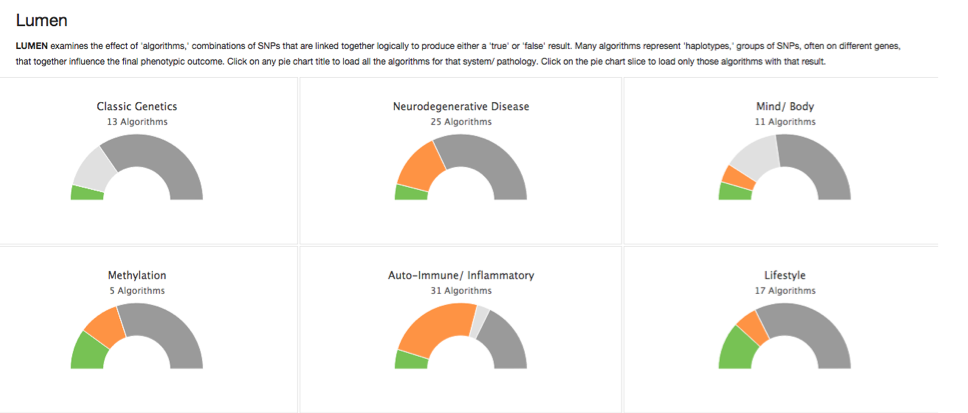
Algorithms are perhaps the most significant and flexible aspect of Opus 23 data. They are usually the easiest result for the non-medical person to understand, because their conclusions are usually simplified statements in everyday language. Algorithms are processed by the LUMEN app in Opus 23 Pro. LUMEN is one of […]