There has been an explosion of interest in the microbiome. Outfits such as uBiome has made it relatively inexpensive and easy to have your microbiome profiled. These services extract the bacterial DNA out of the sample and identify each of the bacteria that the DNA came from.
There appear to be some limitations with the technology. I’ve been told by sources whom I consider informed that uBiome is not that accurate the deeper into phylogeny. Genera data may only be 40-60% reliable by some estimates. So while the major distinctions such as phylum and class may be reliable, drilling down to the precise distribution amounts of particular species may not be so helpful.
Nonetheless there may be some advantages to importing microbiome data into Opus 23 Pro. From a research perspective we’d have the benefit of cross-comparing genomic data with microbiome data, and the ability to perhaps correlate dietary changes based on genomic analysis with progressive changes in sequential microbiome samples.
One advantage is the ease of working with uBiome raw data. The most basic raw data you can download is a simple JSON data file that usually runs about 30K in size, so we’re not talking about any sort of server stress. This data is straightforward enough to parse. The image above was generated out of some basic uBiome data and ported to a visualization script (D3.js) to produce a sunburst information distribution.
The same data ported to a dendrogram based on taxonomic distribution:
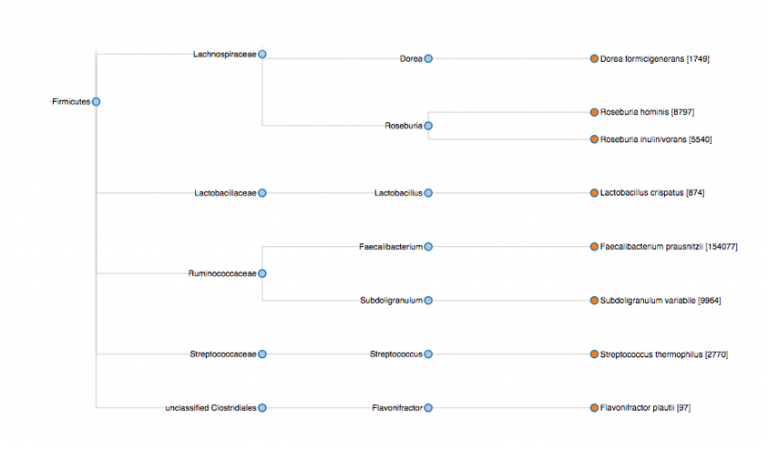
Although this feature will probably not ship with Opus 23 Pro when it hits the pavement in January, I’ll probably add this microbiome tracking ability sometime shortly afterwards.
